Previous articles in this series focused on handling very large text files. At some point, you may be interested in searching for a specific pattern in those large files. Manually searching through a large text file is a non-starter, so leveraging the incredible tools of the developer’s trade is where we turn for help in today’s article.
Regular expressions
Regular expressions (Regex) are built for this task. They are encoded text strings focused on matching and manipulating patterns in the text. They were born into our world in the 1970s. They are extremely useful and considered the key to powerful text processing.
To be more precise, a regular expression is a string that contains a combination of normal characters and special metacharacters. The normal characters are present to match themselves. On the other hand, the metacharacters represent ideas such as quantity and location of characters.
Regex is a language in and of itself, with special syntax and instructions to implement. It can be used with programming languages, like Ruby, to accomplish different tasks, such as:
- Finding text that matches the pattern within a larger text (i.e. our very large text file)
- Replacing the text matching the pattern with other text
- Searching for a file containing the text ant for example, but not if that text is at the end of the word (i.e. want)
These are just a few of the example tasks that are possible. Such tasks can range in complexity from a simple text editor’s search command to a powerful text processing language.
The bottom line is that you, as a Ruby programmer, will be armed with a very versatile tool that can be used to perform all sorts of text processing tasks.
The example today will focus on the main types of tasks regex performs: Search (locate text) and Replace (edit located text).
Searching with Regex
Regex comes in handy when searching text, especially when the text is not a straightforward match. As we mentioned above, you may be interested in finding the text ==ant==. This is simple. But when the location of ==ant== matters, such that you want ant but not want, regex is perfect.
Replacing with Regex
Replacing in regex is a power on itself to be added to the search capability of regex. An example when replacing may be needed is when you want to replace extracted (searched) URLs with clickable URLs, that is, a URL having the HTML href attribute.
A taste of Regex
Let’s do some simple examples with regex to warm up. You can use these tables as a reference for some of the metacharacters we’ll use. Also, as a way to test your regex, use Rubular, an online Ruby-based regular expression editor for testing regular expressions.
Example #1
Let us take this regex for instance:
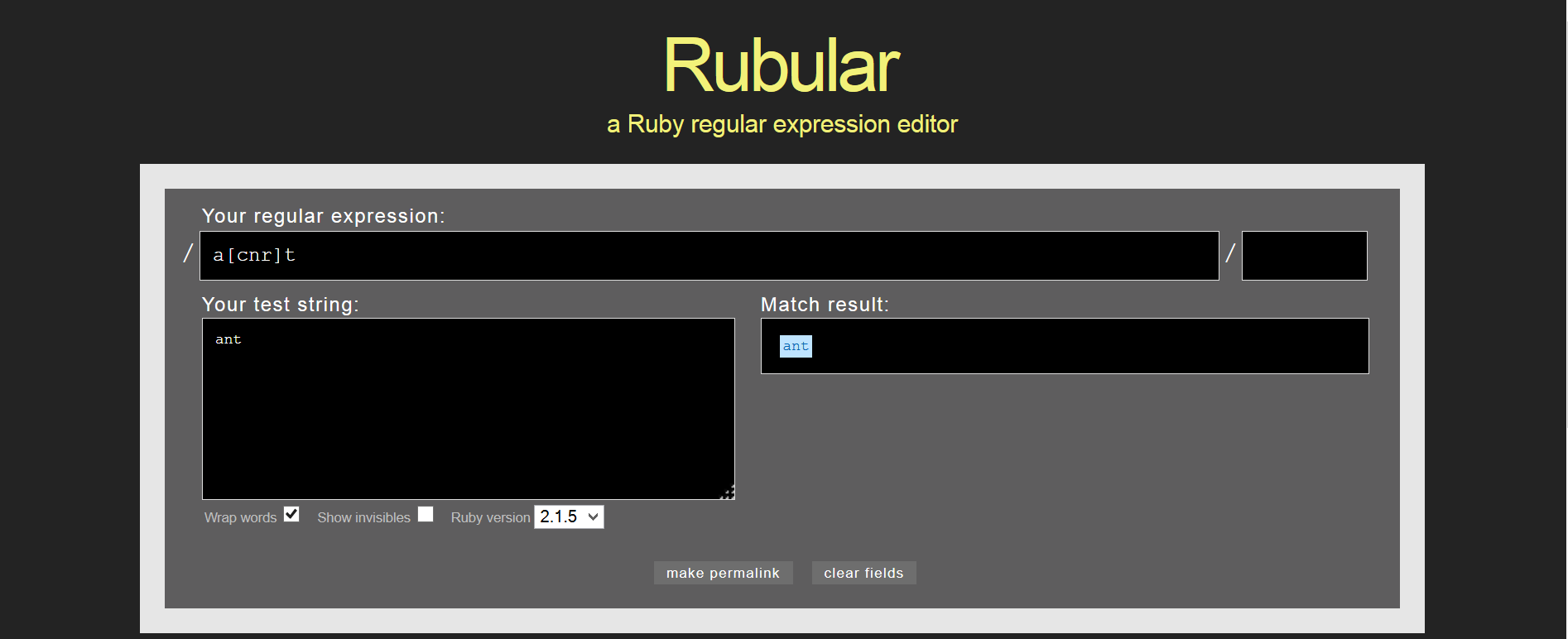
/a[cnr]t/This regex is telling us to find a pattern where the text starts with the letter a, ends with the letter t, and the middle letter is one of c, n, r.
So, the matching words, in this case, are act, ant, and art.
Testing this in Rubular looks something like this:
Example #2
Let’s take this regex for instance:
^HereThis regex matches any string that starts with Here. The circumflex accent (^) metacharacter forces the pattern to start with what follows, in this case Here. For instance, if you had the string Here is the book_, it is matched using the above regex.
Example #3
book.$This regex will match a string that ends with book.. The dollar sign metacharacter forces the pattern to end with what precedes it. For instance, the string Here is the book. matches this regex.
Example #4
bookThis matches a string that has the word book in it. For instance, this string will be matched using this regex The book in on the table.
Example #5
^[A-Z][a-z]+\s[0-9]*Whoa! What’s that??? Don’t worry, this regex looks scary, but is not that complex. What this regex is telling us is to find the string which begins with an uppercase letter ([A-Z]), followed by one or more lowercase letters ([a-z]+), followed by a space (\s), and ending with one or more numerical digits ([0-9]). Brackets ([]) denote a range, meaning, match anything within that range. The + after indicates one or more matches of the immediately preceding expression.
An example of a matching string for this expression is Ali 2015.
Of course, there are many many ways to write regex, and these are just some examples.
Hunting for the Gene Sequence
A gene consists of a long combination of four different nucleotide bases, provided that we have thousands of genes. The four nucleotides are:
- A (adenine)
- C (cytosine)
- G (guanine)
- T (thymine)
Different combinations of those nucleotides give us different characteristics.
In the large file we have (from the previous bits of this series), you will notice a long list of gene sequences. Here is a very tiny snapshot of what is included in the file!
AGGCTTCGATGCCCCTCCACACCCTCTTGATCTTCCCTGTGATGTCATCT
GGAGCCCTGCTGCTTGCGGTGGCCTATAAAGCCTCCTAGTCTGGCTCCAA
GGCCTGGCAGAGTCTTTCCCAGGGAAAGCTACAAGCAGCAAACAGTCTGC
ATGGGTCATCCCCTTCACTCCCAGCTCAGAGCCCAGGCCAGGGGCCCCCA
AGAAAGGCTCTGGTGGAGAACCTGTGCATGAAGGCTGTCAACCAGTCCAT
AGGCAAGCCTGGCTGCCTCCAGCTGGGTCGACAGACAGGGGCTGGAGAAG
GGGAGAAGAGGAAAGTGAGGTTGCCTGCCCTGTCTCCTACCTGAGGCTGA
GGAAGGAGAAGGGGATGCACTGTTGGGGAGGCAGCTGTAACTCAAAGCCT
TAGCCTCTGTTCCCACGAAGGCAGGGCCATCAGGCACCAAAGGGATTCTG
CCAGCATAGTGCTCCTGGACCAGTGATACACCCGGCACCCTGTCCTGGAC
ACGCTGTTGGCCTGGATCTGAGCCCTGGTGGAGGTCAAAGCCACCTTTGG
TTCTGCCATTGCTGCTGTGTGGAAGTTCACTCCTGCCTTTTCCTTTCCCT
AGAGCCTCCACCACCCCGAGATCACATTTCTCACTGCCTTTTGTCTGCCC
AGTTTCACCAGAAGTAGGCCTCTTCCTGACAGGCAGCTGCACCACTGCCT
GGCGCTGTGCCCTTCCTTTGCTCTGCCCGCTGGAGACGGTGTTTGTCATG
GGCCTGGTCTGCAGGGATCCTGCTACAAAGGTGAAACCCAGGAGAGTGTG
GAGTCCAGAGTGTTGCCAGGACCCAGGCACAGGCATTAGTGCCCGTTGGA
GAAAACAGGGGAATCCCGAAGAAATGGTGGGTCCTGGCCATCCGTGAGAT
CTTCCCAGGGCAGCTCCCCTCTGTGGAATCCAATCTGTCTTCCATCCTGC
GTGGCCGAGGGCCAGGCTTCTCACTGGGCCTCTGCAGGAGGCTGCCATTT
GTCCTGCCCACCTTCTTAGAAGCGAGACGGAGCAGACCCATCTGCTACTG
CCCTTTCTATAATAACTAAAGTTAGCTGCCCTGGACTATTCACCCCCTAG
TCTCAATTTAAGAAGATCCCCATGGCCACAGGGCCCCTGCCTGGGGGCTT
GTCACCTCCCCCACCTTCTTCCTGAGTCATTCCTGCAGCCTTGCTCCCTA
ACCTGCCCCACAGCCTTGCCTGGATTTCTATCTCCCTGGCTTGGTGCCAG
TTCCTCCAAGTCGATGGCACCTCCCTCCCTCTCAACCACTTGAGCAAACT
CCAAGACATCTTCTACCCCAACACCAGCAATTGTGCCAAGGGCCATTAGG
CTCTCAGCATGACTATTTTTAGAGACCCCGTGTCTGTCACTGAAACCTTT
TTTGTGGGAGACTATTCCTCCCATCTGCAACAGCTGCCCCTGCTGACTGC
CCTTCTCTCCTCCCTCTCATCCCAGAGAAACAGGTCAGCTGGGAGCTTCT
GCCCCCACTGCCTAGGGACCAACAGGGGCAGGAGGCAGTCACTGACCCCG
AGACGTTTGCATCCTGCACAGCTAGAGATCCTTTATTAAAAGCACACTGT
TGGTTTCTGCTCAGTTCTTTATTGATTGGTGTGCCGTTTTCTCTGGAAGC
CTCTTAAGAACACAGTGGCGCAGGCTGGGTGGAGCCGTCCCCCCATGGAG
CACAGGCAGACAGAAGTCCCCGCCCCAGCTGTGTGGCCTCAAGCCAGCCT
TCCGCTCCTTGAAGCTGGTCTCCACACAGTGCTGGTTCCGTCACCCCCTC
CCAAGGAAGTAGGTCTGAGCAGCTTGTCCTGGCTGTGTCCATGTCAGAGC
AACGGCCCAAGTCTGGGTCTGGGGGGGAAGGTGTCATGGAGCCCCCTACG
ATTCCCAGTCGTCCTCGTCCTCCTCTGCCTGTGGCTGCTGCGGTGGCGGC
AGAGGAGGGATGGAGTCTGACACGCGGGCAAAGGCTCCTCCGGGCCCCTC
ACCAGCCCCAGGTCCTTTCCCAGAGATGCCTGGAGGGAAAAGGCTGAGTG
AGGGTGGTTGGTGGGAAACCCTGGTTCCCCCAGCCCCCGGAGACTTAAAT
ACAGGAAGAAAAAGGCAGGACAGAATTACAAGGTGCTGGCCCAGGGCGGG
CAGCGGCCCTGCCTCCTACCCTTGCGCCTCATGACCAGCTTGTTGAAGAG
ATCCGACATCAAGTGCCCACCTTGGCTCGTGGCTCTCACTGCAACGGGAALet’s say that we want that gene sequence that starts with CTGA and ends with CACT. Between those two patterns, we want either an A, C, or T.
In the tutorial about handling large files, we solved the issue of opening a large file. So, you can now see the content in such a large file, but can you search for out pattern manually? I bet you will have a very difficult time doing so.
Regex to the rescue! This would be a simple task using regular expressions. For such a pattern, we can simply tell Ruby that we want the following to be retrieved:
CTGA(A|C|T)CACTRuby and Regex
Ruby is a very regular expression-friendly language. The Ruby script that matches the gene sequence we want looks like:
puts 'Enter the filename you want to search, and hit ENTER'
filename = gets.chomp
puts 'Enter the regular expression you want to match, and hit ENTER'
regular_expression = gets.chomp
input_file = File.open(filename,'r')
output_file = 'result.txt'
output_file = File.open(output_file,'w')
input_file.each_line do |regex|
if (regex =~ /#{regular_expression}/)
output_file.print regex
end
end
exitThis statement /#{regular_expression}/ creates the regex on the fly from the contents of regular_expression, which we grab from the user. =~ is Ruby’s pattern matching operator, described by the Ruby docs as:
=~ is Ruby’s basic pattern-matching operator. When one operand is a regular expression and the other is a string then the regular expression is used as a pattern to match against the string. (This operator is equivalently defined by Regexp and String so the order of String and Regexp do not matter. Other classes may have different implementations of =~.) If a match is found, the operator returns index of first match in string, otherwise it returns nil.
Running the Script
The snapshot below shows the commands used to run the script:
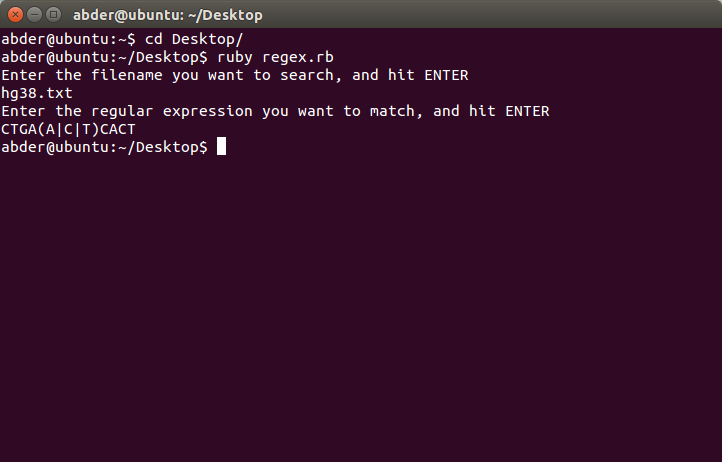
When the program runs successfully, a file called result.txt is created with lines where a pattern match occurs are listed. You can view my version of result.txt here.
More Regex…
If you want to go more deeper in regular expressions, I’d recommended the book Mastering Regular Expressions by O’Reilly.
Happy Rubying and regexing!
 Abder-Rahman Ali
Abder-Rahman AliDoctor and author focussed on leveraging machine/deep learning and image processing in medical image analysis.