There I was, visiting the Sequence and Annotation Downloads page on the UCSC Genome Bioinformatics website. That page contains links to sequences and annotation data downloads for the genome assemblies that are featured in the UCSC Genome Browser. There were so many files to choose from, but I was interested in downloading the following file in the assembly of the human genome data set:
hg38.fa.gz – “Soft-masked” assembly sequence in one file. Repeats from RepeatMasker and Tandem Repeats Finder (with period of 12 or less) are shown in lower case; non-repeating sequence is shown in upper case.
Guess what? That file is greater than 3GB in size! No worries, you may say. Text editors today can handle massive files, right?? I am using Windows, so we’re talking about Notepad, WordPad, and Microsoft Office Word, just to name a few.
Well, it seems we have overestimated the abilities of these editors. When I tried the text editors mentioned above, they screamed in agony. Check it out:
Notepad
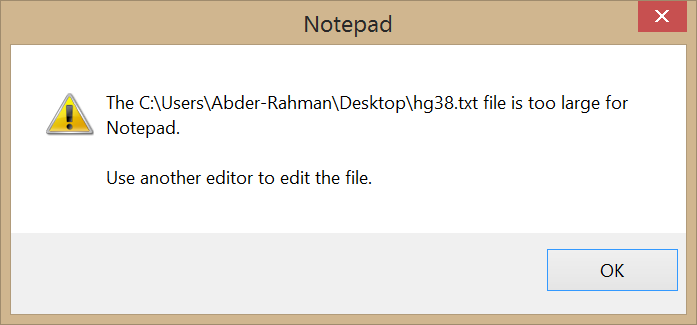
WordPad
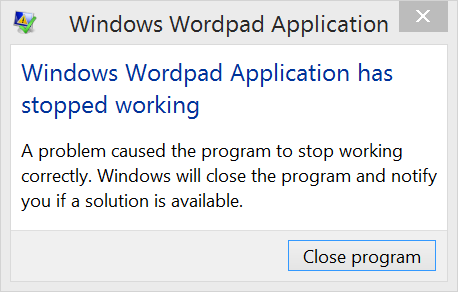
Microsoft Office Word
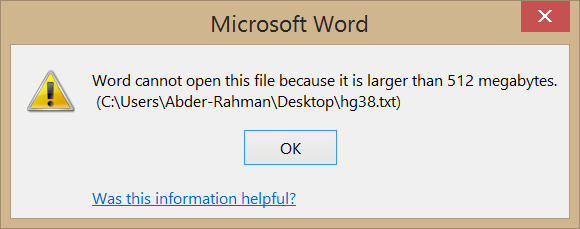
Yikes.
Our goal here is to take a quick look at parts of that large file. In future tutorials of this series, we will see how to use Ruby to navigate through such large files.
Before moving forward with this tutorial, let’s go over some terminology that will make our life easier.
Terminology
Genome (Taken from the Genetics Home Reference)
Based on the Genetics Home Reference, a genome is an organism’s complete set of DNA, including all of its genes. Each genome in turn contains all the information needed to build and maintain that organism. In humans, a copy of the entire genome -more than 3 billion DNA base pairs- is contained in all cells that have a nucleus.
Genome sequencing
As the Genome News Network states, genome sequencing is the process of figuring out the order of DNA nucleotides, or basis, in a genome. That is, the orders of As, Cs, Gs, and Ts that make up an organism’s DNA. As stated above, the human genome is made up of over 3 billion of these genetic letters.
Genome assembly
The genome assembly as Ensembl states, is the genome sequence produced after chromosomes have been fragmented, those fragments have been sequenced, and the resulting sequences have been put back together.
But you already know all that, right? ;)
Obtaining the File
In this section, let’s grab that large file mentioned at the beginning of this tutorial.
The good folks at UCSC Genome Bioinformatics are generous enough to supply a kind of readme that describes each of these gigantic files and their content. The file we want to download is hg38.fa.gz (be careful, the file size is 938 MB).
After you download the file, go ahead and extract it. You can use WinRAR or 7-Zip or tar. After you extract the file, you’ll have a file called hg38.fa. Go ahead and rename it to hg38.txt, so we have a txt file instead of an fa file. As mentioned at the beginning of this tutorial, using Notepad, WordPad, and Microsoft Office Word, the file can’t be opened due to its large size.
A Journey of a Thousand Lines
Let’s see what it takes to use Ruby to extract 1000 lines of that large file at a time. Of course, you can extract as many lines as you like. The first thing we want to do here, is ask the user to provide the name of the file to snoop. For this, do the following:
puts 'Enter the name of the file'After asking the user for the file name, we need to read that file name and store it in a variable. This can be achieved in Ruby as follows:
file_name = gets.chompLet’s analyze what is happening here a bit. gets is a method used to get the next line from standard input as a string, up until the return. chomp is used to return the string without the terminating line return \n. If we don’t chomp the string, the program returned the following error:
snoop.rb:4:in `initialize': Invalid argument - hg38.txt (Errno::EINVAL)
from snoop.rb:4:in `open'
from snoop.rb:4:in `<class:XYZ>'
from snoop.rb:1:in `<main>'From the error, it seems that Ruby was reading the following filename hg38.txt\n instead of hg38.txt.
After reading the file name, we now want to open() the file:
input_file = File.open(file_name,'r')r here means that we are opening the file in read mode.
Our script will write the extracted lines to another file, which those wimpy text editors can handle. In this case, File.open(), with a w (write) mode:
output_file = File.open('output.txt','w')We will be reading line-by-line from the input file. The Ruby method that will aid us in this process is readline():
read_line = input_file.readlinewrite() is required in order to write the read lines to the output file. Yes, I mentioned lines here (i.e. with an “s”). But, we are using readline() that reads only one line at a time. To start, a for will limit our reading to 1000 lines.
At this point, let’s see how the Ruby script performing the operations discussed in the previous section would look like.
class Snoop
puts 'Enter the file name you want to work with'
file_name = gets.chomp
input_file = File.open(file_name,'r')
output_file = File.open('output.txt','w')
for i in 1..1000
read_line = input_file.readline
output_file.write(read_line)
end
endRunning the program
To run the program, I used the Command Prompt with Ruby on a Windows 8.1 machine, as follows:
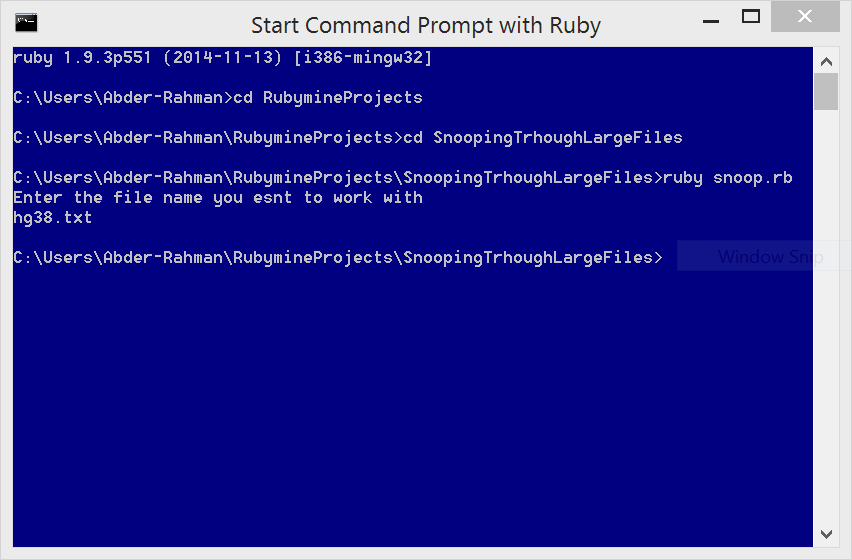
The result of the program (i.e. output.txt), can be downloaded from here.
Why Do We Need This?
As a Ruby programmer, you may notice that the program is simple. Let’s not forget that, since this series is geared towards applying Ruby to medicine related topics, we could expect non-programmers rolling in (i.e. scientists, biologists, researchers) looking for a solution to their issues.
For a programmer, maybe the content in the large file we have been working with doesn’t make much sense. It does, however, matter to the specialist working with the file.
Specialists are frequently faced with such large files and, as we have seen, the text editors that we use often are not able to deal with such files. The point of the program presented in this tutorial is to be simple (Ruby comes to play) for scientists, researchers, etc. to understand and utilize. The techniques used in this article get the specialist exactly what is needed: A smaller file with relevant output that can be opened in a standard text editor. With the technology hurdles out of the way, the specialist can now focus on improving lives and, subsequently, the world.
OK, maybe that is a bit over-the-top, but the core of it is true. This series not only bring medical concerns to Ruby, but also (and, perhaps, more importantly) brings Ruby to the medical field.
 Abder-Rahman Ali
Abder-Rahman AliDoctor and author focussed on leveraging machine/deep learning and image processing in medical image analysis.